Imaging Data
Accuracy of stereotactic planning greatly depends on the available diagnostic sequences, imaging parameters of those. DICOM/NIFTI header contains these data in readable form together with reference to imaging volume and can be extracted and displayed in a parsing window. Surgical planning is based on 3D volumetric model (assuming equal slice distances) and in case of any difference larger than a preset error limit the diagnostic sequence can not be imported. Sequences with special conversion (like Siemens well-known JPEG lossless format for MR) can be imported for surgical planning. The header of DICOM/NIFTI should contain all parameters according to scanner’s setting. Any conversion by third-party software could produce errors or clear important parameters therefore not recommended. CranioPass sorts the imported images according to scanner table automatically and verifies their 3D usability. Using patient database one can identify not only diagnostic volumes but archive studies as well, with list of important elements (CT-MR fusion parameters, type and 3D path of electrodes, target coordinates in stereotactic space).
Modalities/Data Formats
Three diagnostical sequences are distinguished by CranioPass:
- Reference slice sequence (usually CT modality and available just before surgery), which contains markers for registration of stereotactic space. These markers usually are intercepts between the marker lines (on plates fixed to skull) with CT slices (in different arrangements depending on the used type of head-frame. This imaging sequence is made just before surgery;
- Fusion slice sequence (usually MR modality), which has been scanned days before surgery and doesn’t have markers. It gives information on soft tissue. As volumetric model, it should be registered to reference sequence;
- Post-operative (PostOp) diagnostic sequence for testing success of surgery (after registration to reference volume). Fusion slice sequence can be replaced by PostOp diagnostics and, after image fusion, the path of intervention and target positions can be displayed on it.
CranioPass is able to import CT (computer tomographic) and MR (magnetic resonance) imaging sequences as standard DICOM images. Both MR T1 and T2 formats are acceptable. In addition, NIFTI-1 formats (http://nifti.nimh.nih.gov/nifti-1/documentation) is also readable. NIFTI standard supports usage of many, new MR diagnostic data types (like functional MR imaging). Both uncompressed NIFTI (.nii) and compressed (.nii.gz) formats are readable for planning. MR sequence in Siemens “lossless” JPEG format is also readable.
Selection of Image Sequence (Automatic Data Parsing)
First module of CranioPass (“Patient Data”) is able to create a patients’ database. This database stores and recalls in a user-friendly way the parameters of diagnostic volumes and elements of earlier surgical plans. The surgical plan can be archived into study file (.vts) which later can be used for verification of the surgical procedure or follow-up consultation. CranioPass, during the startup procedure of the program, automatically builds up the database from a list of patients located in a root folder. This root folder is initialized by the user and can be modified at any time from the program. The program with this database is able to build up complex patient/diagnostics sequence references so that from the header of selected DICOM/NIFTI/study file the important parameters are displayed in the informational window. This helps communication between the surgeon and diagnostic laboratory that especially important in case of large mass imaging data. The informational window gives info to surgeon for selecting appropriate image sequence and during data parsing from .vts study the surgical plan is made more transparent. The folder for patient may contain: (1) Reference sequence, (2) Fusion sequences, (3) Archive studies (Surgical plans in compressed vts files). NIFTI (.nii or .nii.gz) images are readable, as Reference or Fusion sequences. Special diffusion MR (DWI Diffusion Weighted Image) sequences are recognized and their parameters can be listed in the informational (Data Parsing) window.
Image 4. Upload patients’ database from patients’ data folders located in root folder which can be set from the application. CranioPass automatically extracts most important parameters of DICOM/NIFTI diagnostic sequences and elements of surgical planning as stored in archive studies (.vts). The diagnostic sequences are identified by fie paths extracted from .vts type archive file. The results are displayed in informational window making easier the recognition of diagnostic environment and reconstruction of different planning settings.
Visualization in CranioPass
As a first step, the program always imports the reference sequence and sets, accordingly, the size of reference voxel. Further on, the next sequences (i.e. MR modality to image fusion, PostOp sequence) will be interpolated to this reference voxel size in 3D. Surface reconstruction is based always on reference volume. The replacement of reference series is possible only before importing fusion sequence. The fusion or PostOp series are replaceable but the registration between the overwritten data and reference should be repeated for correct visualization.
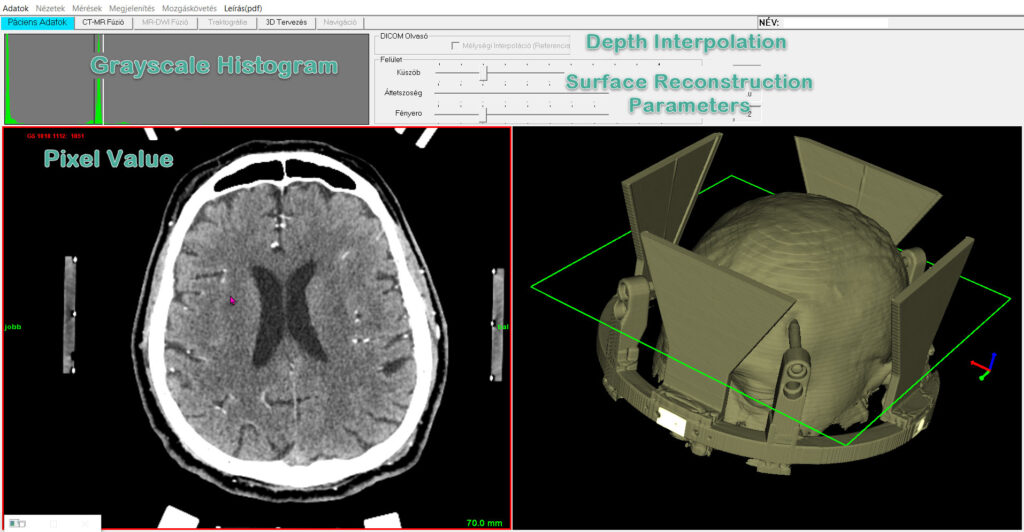
Visualization settings of reference volume and quality of surface reconstruction can be adjusted in the first panel (“Patient Data”). The program is able to import reference series under “Depth Interpolation”. If this option is enabled, the program automatically sets the distance between slices to same value as x-y pixel size that might increase the number of slices after interpolation. This way the resolution and quality of surface reconstruction can be increased, but at the same time the need of memory is getting higher during diagnostic data processing.
Grayscale Histogram
The displaying quality of reference volume can be adjusted by grayscale histogram (Image 5). Black/white limits are changeable separately or with moving together. The results of adjustment are validated for diagnostic background in all panels (Patient Data, CT-MR Fusion, 3D Planning).
Surface Reconstruction
Quality of surface reconstruction can be modified by means of 3 parameters:
- Grayscale threshold (bone threshold this time, other segmentation is not possible in this version);
- Transparency ratio for opaque view;
- Lighting intensity.
Interactive adjustments are needed i.e. change in transparency implements adjusting of light intensity to keep surface visibility.
Image 5. Parameter selection and visualization in “Patient Data” panel. Green frame depicts the actual slice position in surface view (right).
Resampling of Image Sequence for Reference
CranioPass can be used for fixed and user defined resampling, into any orientation even in real time, of reference volume. With fixed procedure the original sequence can be reflected across any of orthographic planes and the slice order can be reversed. Furthermore, the reference volume can be resampled about XYZ axes with rotation center placed anywhere in 3D. The volumetric model can be rotated about axes assigned in axial, frontal and sagittal planes in an interactive way and the cumulative result is shown by resampled axial volume (second row, right window). By unselecting this resampling mode in menu, the program updates the reference sequence and the surgical planning is performed on the resampled reference background. The calculation updates the reference view in all panels and the surface view is refreshed accordingly. If archived, the archive study (.vts) stores the rotation angles and rotation center. The reformatted axial output (second row right) can be reseted by double click which clears rotation angles and restores the original imaging sequence.
Alignment to Patient’s Anatomy (AC-PC)
Experienced user can set easily any slice orientation according to needed anatomic orientation. Image 6 illustrates settings for matching AC-PC orientation performed by rotations in 3 orthographic views.
Image 6. Reformatting reference CT sequence according to the plane crossing anatomic points (AC-PC). Center of rotation and the rotation angles are set arbitrarily in 3D by selecting 3 axes. Interpolated (with rotation angles) and resampled CT slice and reconstructed surface are shown on the right side of 2 lower rows.